Characterizing Expression Patterns of LexA and LexAop Lines for the Development of a Drosophila Cancer Model
Published · Updated

A report written by Nora Solheim
Abstract
Drosophila melanogaster (the fruit fly) is a very well-suited model organism within the genetic fields of biology. It has a short generation time and a simpler genome than that of humans, providing a good opportunity to control gene expression within the organism. The genome of Drosophila is closely related to that of humans where 75% of disease-related genes overlap. Cancer is a well-known disease in which some of the cells in the body grow uncontrollably, and per today there is no cure. Malignant tumour growth and spread are the cause of mortality in more than 90% of all cancer patients, and the mechanisms of what causes malignancy are poorly understood. This experiment is one of many steps necessary to obtain a greater understanding of the communication between cancer cells and healthy surrounding tissue cells. Drosophila offers a system where a tumour can be induced in a restricted region using RET/Stit combined with LexA for controlling expression patterns. The surrounding neighboring cells can also be controlled by using a second binary system which Drosophila provides. We were provided with a selection of different LexA driver lines and have through this experiment identified their expression sites, finding suitable LexA drivers which can be used to compare RET/Stit expression to tumour development. Two of the lines were exposed to Stit to see which effect this might have. The results indicate that Stit promotes the spreading of cells as the expression sites altered from the control samples.
Introduction
The body size of an organism is an important trait developed over time to adapt to a specific environment (Layalle et al., 2008). Cellular and organismal growth in animals depends on two factors: the rate of growth, and the duration of the growth period. Both these factors are regulated and modulated by environmental factors such as nutritional and hormonal cues (Layalle et al., 2008; O’Farrell et al., 2013). Within many species, the insulin/IGF family plays an important role in setting the growth rate as it is one of the factors connecting nutrition intake and growth (Layalle et al., 2008). In all eukaryotes studied, the conserved protein complex target of rapamycin complex 1 (TORC1), is the link between the insulin/IGF family obtained via nutritional cues and the regulation of cellular growth and proliferation (O’Farrell et al., 2013).
The TORC1 system is initiated by the binding of insulin to the insulin receptor (InR) activating the insulin substrate (IRS), which in turn activates a downstream signalling process. This signalling process includes phosphatidylinositol-kinase class 1 (PI3K-1), which in turn phosphorylates and activates the protein kinase Akt, promoting TORC1 activation (O’Farrell et al., 2013; Schmelzle & Hall, 2000). Recent research has found that the proto-oncogene RET (Rearranged During Transformation) has an impact on the TORC1 system, resulting in cell and tissue overgrowth (O’Farrell et al., 2013). The RET proto-oncogene encodes a receptor tyrosine kinase which is expressed in tumours and tissues originating from the neural crest (Eng, 1999). Different rearrangements of RET have been detected in several different varieties of human cancers such as lung and thyroid cancer (Takahashi et al., 2020).
Malignant tumour growth and spread are the cause of mortality in more than 90% of all cancer patients. It is a disease in which the abnormal cells divide uncontrollably and can spread to nearby tissues and sometimes other regions and organs of the body through the blood and lymph systems (National Cancer Institute, 2022). A homolog of RET has been identified in Drosophila melanogaster (the fruit fly), first used as a model organism by Thomas Hunt Morgan (Markow, 2015), called Stitcher (Stit). This together with the fact that downstream signalling processes are widely conserved makes Drosophila a good model organism for cancer research. Stit encodes a RET-family receptor tyrosine kinase which is required and activated during epidermal wound healing in Drosophila embryos (O’Farrell et al., 2013; Wang et al., 2009). As a consequence of wound healing, it is also found to promote growth in the Drosophila epithelial imaginal wing discs, where it controls the balanced growth of the dorsal and ventral wing disc compartments. Stitcher is therefore required for optimal growth and activates the TORC1 downstream signalling pathway (O’Farrell et al., 2013; Wang et al., 2009). Both oncogenes RET and Stit show stimulatory effects on cell migration, a process allowing the movement of individual cells or a group of cells from one location to another (Boekhorst & Friedl, 2016).
Understanding the relationship between the microenvironment and the tumour itself requires a complex that enables the induction in one cell while still allowing the surrounding neighbouring cell’s gene expression to be regulated, in other words, two independent binary systems (Lai & Lee, 2006). The combination of the binary systems can therefore be utilised to find out what causes the transition to malignancy, and to what extent the surrounding tissue of a tumour prevents or promotes the migration of aberrant cells. The Drosophila model organism provides such a system, using RET/Stit which promotes tumour combined with LexA for controlling the expression patterns of these cells (Lai & Lee, 2006; Boekhorst & Friedl, 2016).
Drosophila is a great genetic model organism due to a variety of benefits. Drosophila generates a large number of externally laid embryos that are transparent throughout the larval stages of development, have a quick generation time of only 10 days, and is simple and inexpensive to maintain in the lab (Jennings, 2011). Humans and Drosophila share a strong genetic relationship; between the two species, 60% of the genes in general and 75% of disease-associated genes are shared (O’Farrell et al., 2013). Drosophila has a simpler genome, providing a better opportunity to control gene expression. The imaginal wing disc, which becomes the wing of the organism, is easily accessible within the larval stage of development and the genetic expression within this area is easily controllable, making it a suitable organ for characterizing genetic expression (O’Farrell et al., 2013). Drosophila has four chromosomes which make site-specific insertions easier. To ensure that the whole gene (and chromosome) of interest is passed on to the following generation, balancers are used on entire chromosomes to inhibit recombination (Miller et al., 2019).
Drosophila genetics provides the binary expression system GAL4/UAS (Upstream Activation Sequence), consisting of two components GAL4 transcriptional activator which is inserted in a combination of an enhancer. When activated, it expresses GAL4. The UAS promoter is activated in the presence of GAL4, promoting transcription of a gene of choice downstream, in this case being Green Fluorescent Protein (GFP) (Rodriguez et al., 2011). All RNAi (interfering RNA capable of reducing gene expression) lines, as well as the oncogenes RET/Stit, rely on the binary system GAL4/UAS. To knock down the genes within the surrounding cells of the oncogene independent of RET/Stit expression, a separate system is required; one system for controlling the expression of RET/Stit, and one system controlling the expression of the surrounding tissue (Rodriguez et al., 2011). Drosophila provides a second binary system, LexA/Aop. This system has the same mechanism as GAL4/UAS, where LexA binds to and activates the LexAop, the LexA operator (Rodriguez et al., 2011). Further, the two systems are not affected by one another and can work simultaneously within the same tissue, allowing researchers to perform two manipulations of gene expression in vivo (Rodriguez et al., 2011).
The aim of this study is to find suitable LexA driver lines and compare RET/Stit expression to tumour development within the imaginal wing disk of the Drosophila. To achieve this, different genetic lines had to be created by performing numerous crosses of Drosophila flies tagged with different phenotypes to ensure the presence of the desired gene. Our hypothesis is that some of the LexA driver lines will provide suitable and restricted expression patterns, which can later be used to understand the mechanisms of tumour growth and communication within the microenvironment of a tumour. We further hypothesise that the strength of expression will correlate with the penetrance of the tumour phenotype.
Materials & methods
At the University of Bergen's fly lab facility, flies were cultivated in an incubator at 25 °C and with a light regulation that mimicked the daily rhythm of the sun in the flies' natural environment. The light turns on at 9.00 in the morning and off at 9.00 in the evening. Flies were stored in tubes containing a food medium and were flipped1 once a week to a fresh tube of food. All different genetic fly lines were provided by Fergal O’Farrell, associate professor at the University of Bergen (Table 1). Table 1 abbreviations will be used to refer to the genetic lines throughout this paper.
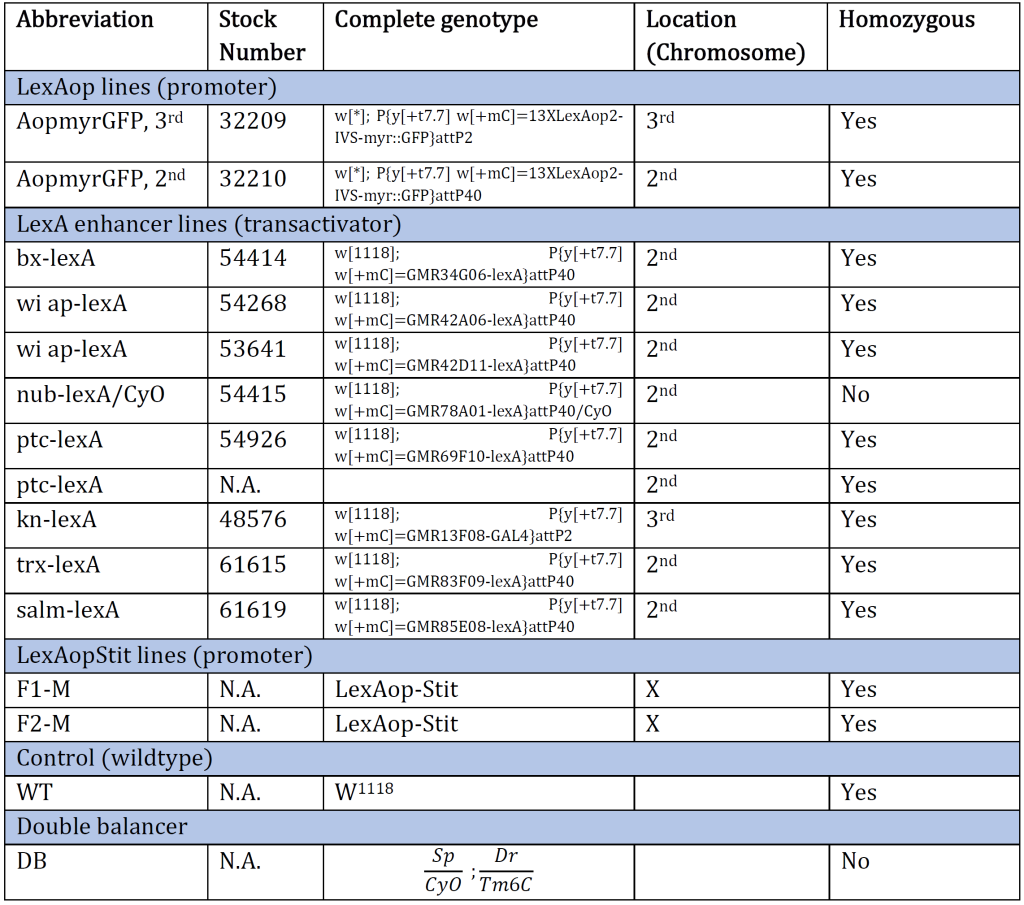
Table 1: An overview of the Drosophila genotype of original genetic lines provided at the beginning of the experiment. The genotype and the corresponding stock number are aligned. The table also provides an abbreviation for each genotype and which chromosome the gene of interest is inserted/present. N.A. indicates information Not Available.
Selection of Flies - Fly Pushing
The balancers had different phenotypic expressions, enabling us to sort out the flies containing the genetic lines of interest with the use of a microscope (Figure 1). The technique used is called fly pushing and refers to the daily sorting of flies. Flies are emptied from their tube onto a CO2-perfused pad, knocking out the flies instantly. A small paintbrush is then used to ‘push’ the flies around to select for or against anatomical characteristics and phenotypic expressions. A Leica light microscope with additional spotlights was used for fly pushing.
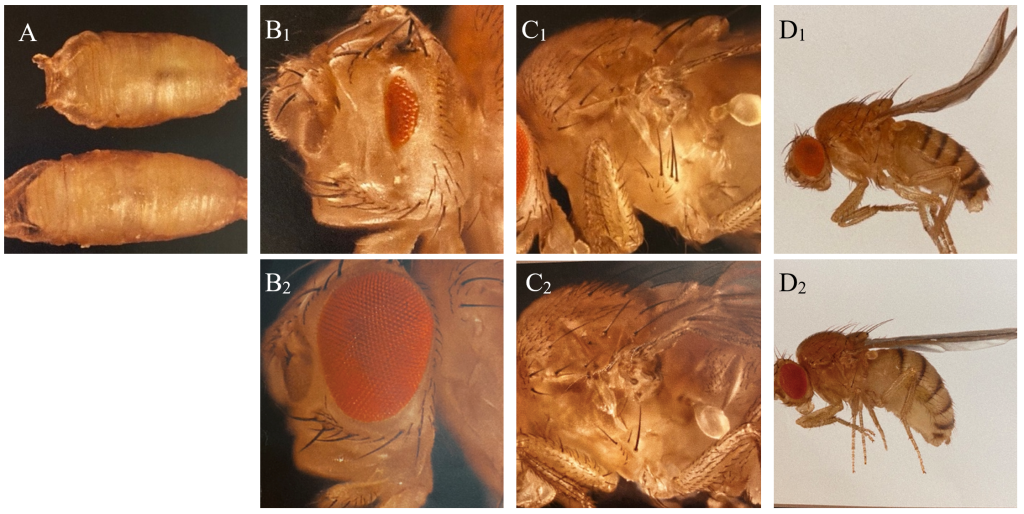
Figure 1: Phenotypic expressions of balancers and corresponding wild type of Drosophila used in the experiment. A: C: Tb (Tm6C), reduction of body size (above), and wildtype (below). B: Dr, reduction of eye (B1) and wildtype (B2). C: Sp, additional hairs behind the first anterior leg of the fly (C1) and wildtype (C2). D: CyO, curly wings (D1) and wildtype (D2).
Setting Crosses
Crosses were set on a Friday, ensuring that the F1 generation started to emerge on a Monday (10 days after fertilization). In order to make a cross, 4-8 virgin females and 2-4 males were required. Virgin females have specific characterisations visible with the use of a microscope. They are more transparent, paler in colour, and possess bloated abdomens. The crosses were flipped to a new tube 4-5 days after the cross had been made, preventing the parent generation and F1 generation to be mixed.
Generation of Double-Balanced Stocks
The goal was to create double-balanced stocks containing a LexA enhancer line on the 2nd chromosome, and LexAopmyr::GFP 3rd chromosome (Figure 2). There was one exception, that of the KnLexA, where the aim was to have both the LexA and LexAopmyr::GFP on the 3rd chromosome. The differentiation was caused due to the position of the KnLexA enhancer on the 3rd chromosome (Figure 2). The goals required a multistep crossing scheme. A short summary of the original genetic lines needed to create each double-balanced stock is included (Figure 2). A slash (/) indicates that the genes are on the same chromosome whereas a semicolon (;) indicates that the genes are on separate chromosomes of Drosophila.
Simultaneously as the multistep process of creating double-balanced stocks started, direct crosses of each LexA promoter line and Aopmyr::GFP were also made (before the enhancers had been balanced). This was done to check if there was a GAL4 expression pattern, shown with GFP-positive cells if present. If the results were negative, meaning no expression pattern, the given LexA enhancer line would not be used further in the experiment.
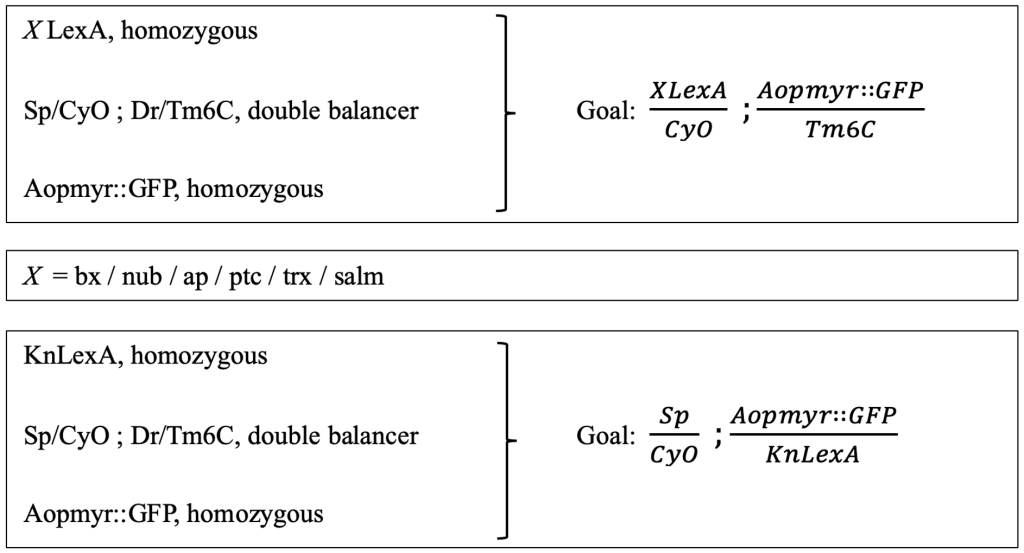
Figure 2: Presentation of the crossing scheme aim for obtaining double balanced stocks. The original lines (left) that were used to create the goal (right). The procedure was consistent for all different LexA enhancer lines; therefore, the X is used in order to indicate each LexA enhancer line. The goal and original lines needed for KnLexA are also shown.
Microscopy and Dissection
The imaginal wing disk of wandering third-instar larvae was removed by dissection and fixed in 500 µl 4% formaldehyde/PBS (Phosphate-Buffered Saline) for 20 minutes in a well plate. Then the disks were rinsed with PBS for 3x1 minute before being fixed in 500µl PBS containing Hoechst 10 µg/ml with a 1:10 000 ratio for 20 minutes. Lastly, the disks were rinsed with PBS.
A drop of glycerol mountant was added to an objective slide by using a pipette. The imaginal wing disks were then transferred to the mountant, and an objective glass was placed on top. The objective slides were labelled immediately afterward with the genotype of the imaginal disks. All objectives were stored in a fridge until microscopy of the disks. The dissection was performed using a surgical needle and forceps. A minimum of three discs showing the same result for each genotypic expression was required before conclusions could be drawn.
Illumination microscopy imaging was performed using a Leica inverted microscope and the LasX Leica software. The disks were illuminated with 420nm and 488nm wavelength light produced by an LED laser light source for the detection of Hoechst and GFP-positive cells/expression, respectively. Following image acquisition samples were compared using ImageJ Fiji.
Double-Balanced Enhancer LexA lines exposed to Stit
The completed double-balanced LexA;AopmyrGFP stocks were crossed with LexAopStit (Stit). The goal was to detect any changes that might occur within the GAL4 expression pattern when being exposed to Stit. At least two samples of each Stit;LexA;AopmyrGFP as well as a control, not crossed to Stit, were made for each completed line following the same procedure as the microscopy and dissection before (section "Microscopy and Dissection"). Due to limited time, only the LexA enhancer lines PtcLexA and ApLexA was exposed to Stit.
Results
Direct Crosses of LexA Promoter lines for GAL4 Expression
The expression patterns of all results are shown with GFP-positive cells, if present, within the imaginal wing disc of Drosophila. The direct cross of different LexA promoter lines for GAL4 expression is presented in Figures 3 – 5. GFP-positive cells were observable in the LexA promoter fragmented genetic lines of Trx, Ap, Ptc, and Nub (Figure 4 – 5), whereas the LexA promoter fragmented genetic lines Bx, Kn, and Salm were GFP-negative (Figure 3). The GFP-positive expression sites varied within the imaginal wing disc among the different genetic lines, however, all lines had some expression within the wing pouch area. Further NubLexA only showed expression in 50% of the imaginal discs dissected due to this line not being homozygous. To confirm GFP-positive cells a tube with larvae was held under a LED light where GFP was visible as a brighter region in the anterior region of the larvae (Figure 6).
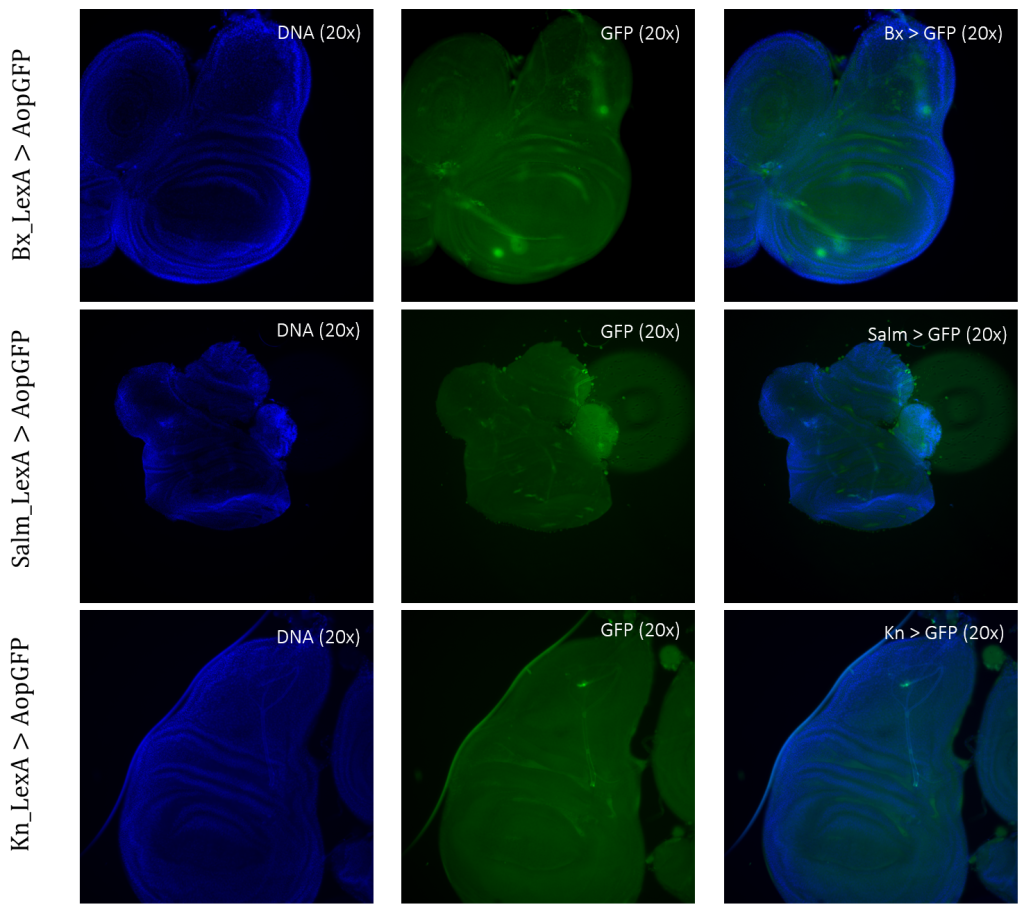
Figure 3: Negative GFP-cells in tissue samples from LexA promoter for GAL4 expression in Drosophila. BxLexA, SalmLexA, KnLexA, showed no expression sites of GFP-positive cells within the imaginal wing disk of Drosophila. The GFP scan shows that there is only background colouring for these driver lines.
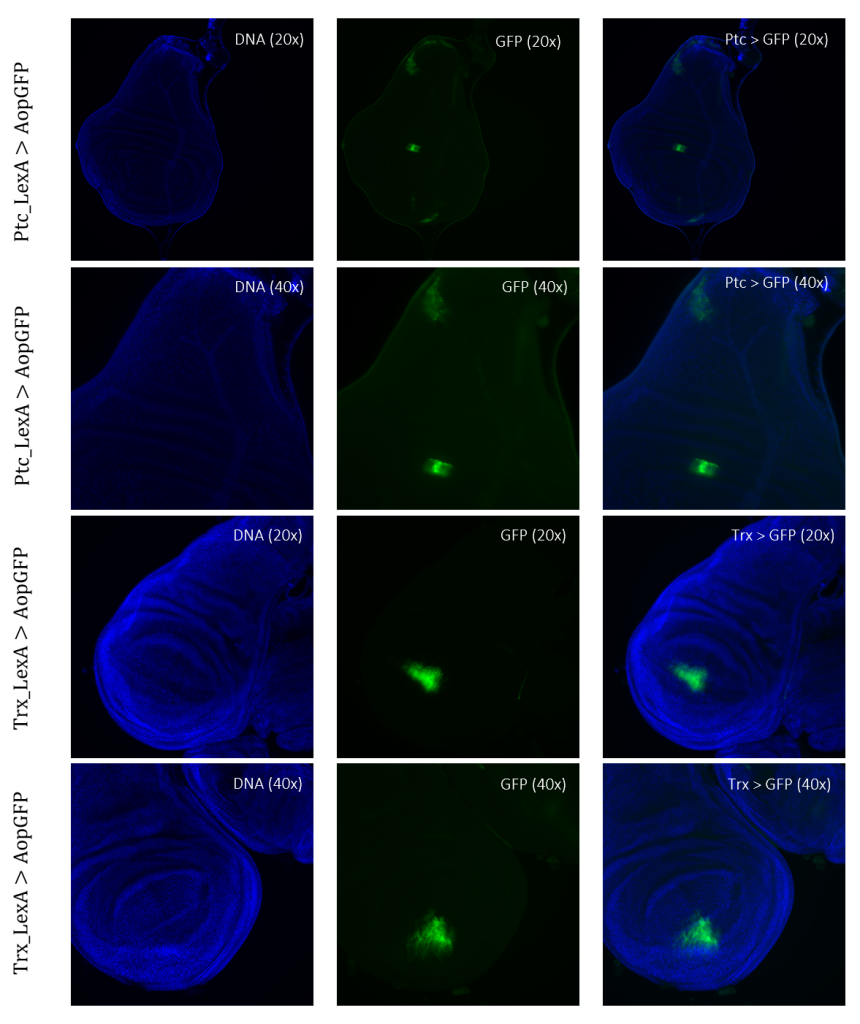
Figure 4: Tissue samples from LexA promoter for GAL4 expression in Drosophila. GFP-positive cells were observable in different regions of the imaginal wing disk of Drosophila for driver lines Ptc(54926)LexA and TrxLexA. PtcLexA showed fragmented expression within the wing disk, including the wing pouch and notch. TrxLexA showed a restricted region of positive GFP cells in the pouch area of the imaginal wing disk.
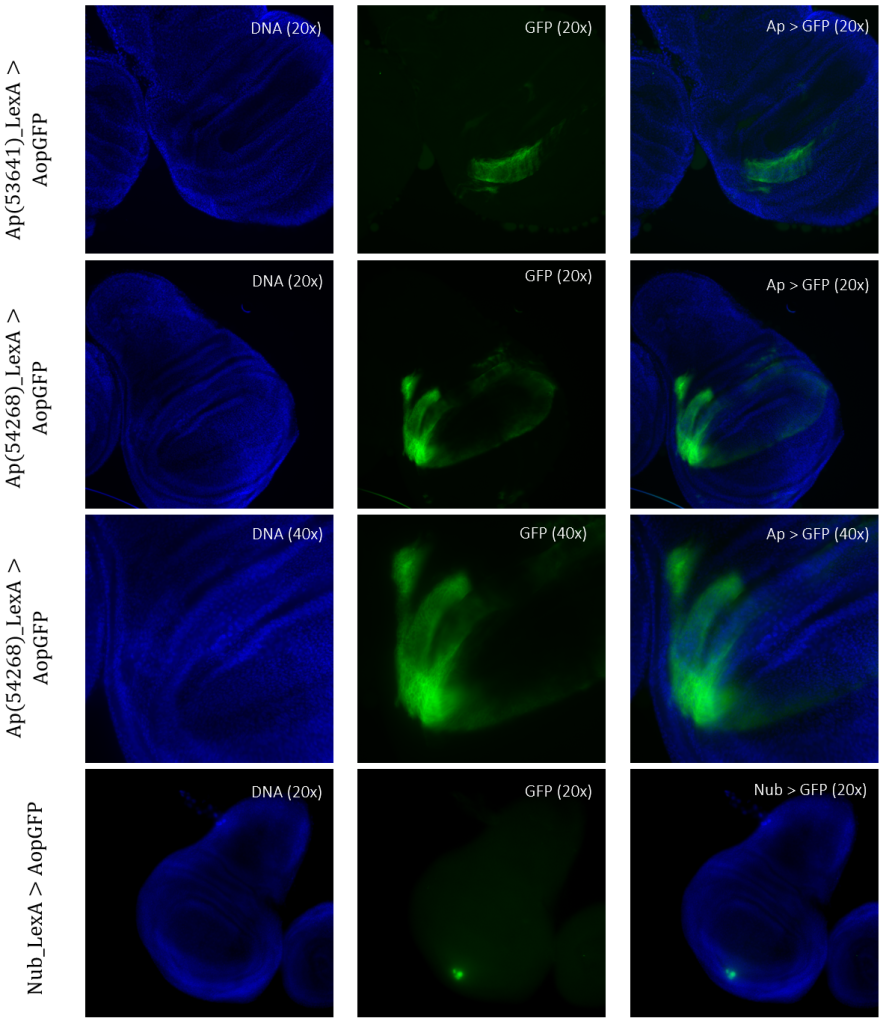
Figure 5: Tissue samples from LexA promoter for GAL4 expression in Drosophila. GFP-positive cells were observable in different regions of the imaginal wing disk of Drosophila for driver lines Ap(53641)LexA, Ap(54268)LexA, and NubLexA. Both ApLexA lines had GFP-positive cells in the dorsal region of the wing pouch, where the expression pattern of Ap(54268) was also present in the wing thorax. NubLexA showed three small fragments of GFP-positive cells within the ventral region of the wing pouch.
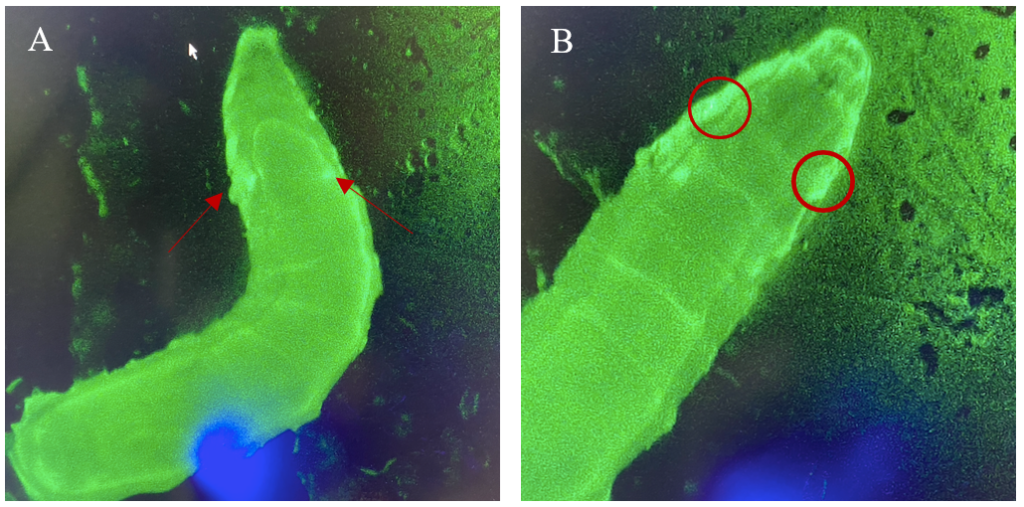
Figure 6: 3rd instar larvae with NubLexA promoter for GAL4 expression in the imaginal wing disc. The GFP-positive region was identified in the anterior region of the larvae as a small circle with a brighter colour on both the left and the right side of the organism. The figure shows one larva to the left (A) and the same larva but at a higher magnification to the right (B). The areas with expressions are highlighted with arrows and circles.
Crosses of Double-Balanced LexA promoter lines for GAL4 Expression when exposed to Stit
Crosses of double-balanced lines Ptc>GFP and Ap>GFP with Stit-transformation of cells showed abnormal expression patterns. Two different positioned Stit-transformations were used, Stit FM1 for Ptc expression (Figure 7) and Stit FM2 for Ap expression (Figure 8). Stit>Ptc>GFP, Stit-transformed cells, showed an elongated expression fragment within the wing pouch of the first sample, less centered than that of the control (Figure 7). The second sample showed more centered fragments similar to that of the control. The region of expressions for both samples were overlapping with that of the control.
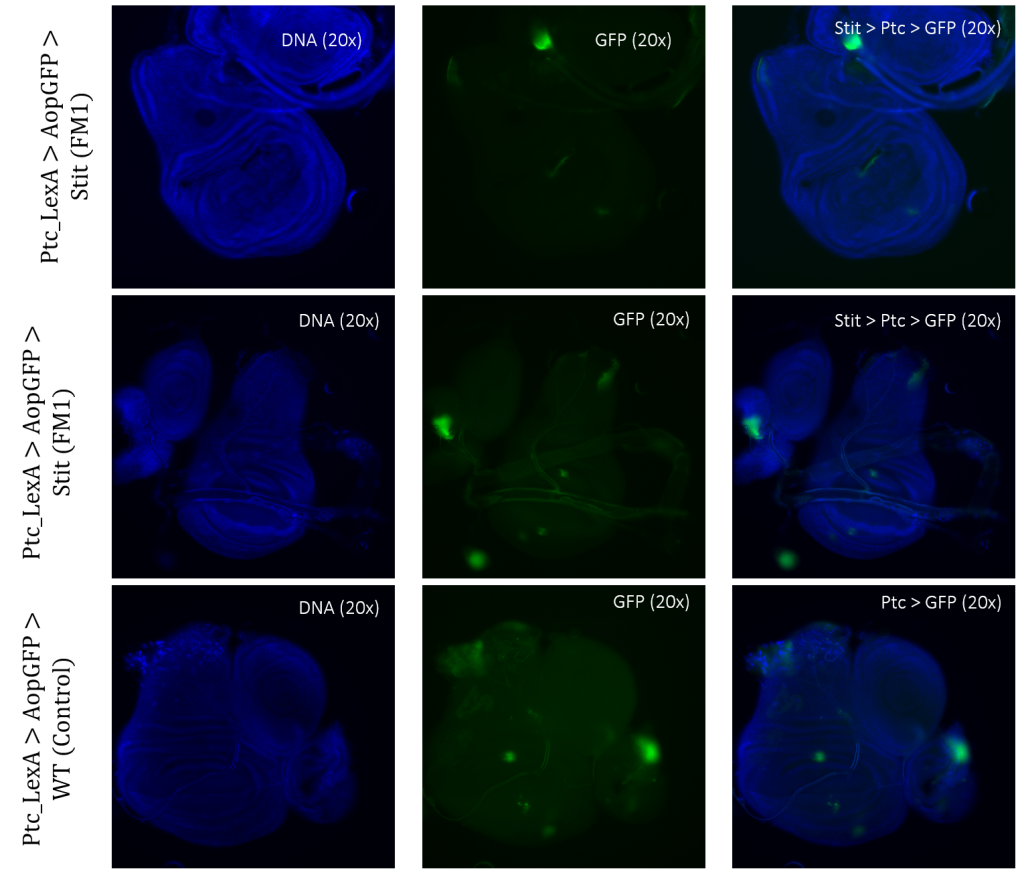
Figure 3.5: Tissue samples from PtcLexA promoter for GAL4 expression when exposed to Stit in Drosophila. Wing disk where PtcLexA>GFP is shown as fragments within the pouch and notch. Stit-transformation of cells (Stit>Ptc>GFP) leads to more diluted fragments of GFP-positive cells. The areas of GFP expression are consistent with that of the control.
Stit>Ap>GFP, Stit-transformed cells, showed a smaller area of GFP-positive cells than that of the control. This result was consistent for both samples of Stit>Ap>GFP (Figure 8). The first sample was disturbed due to the positioning on the objective glass, however, there were still GFP-positive cells present within the wing pouch, which were more elongated than the control. The second sample showed a strongly reduced expression region in the pouch, with two separate fragments instead of one larger fragment.
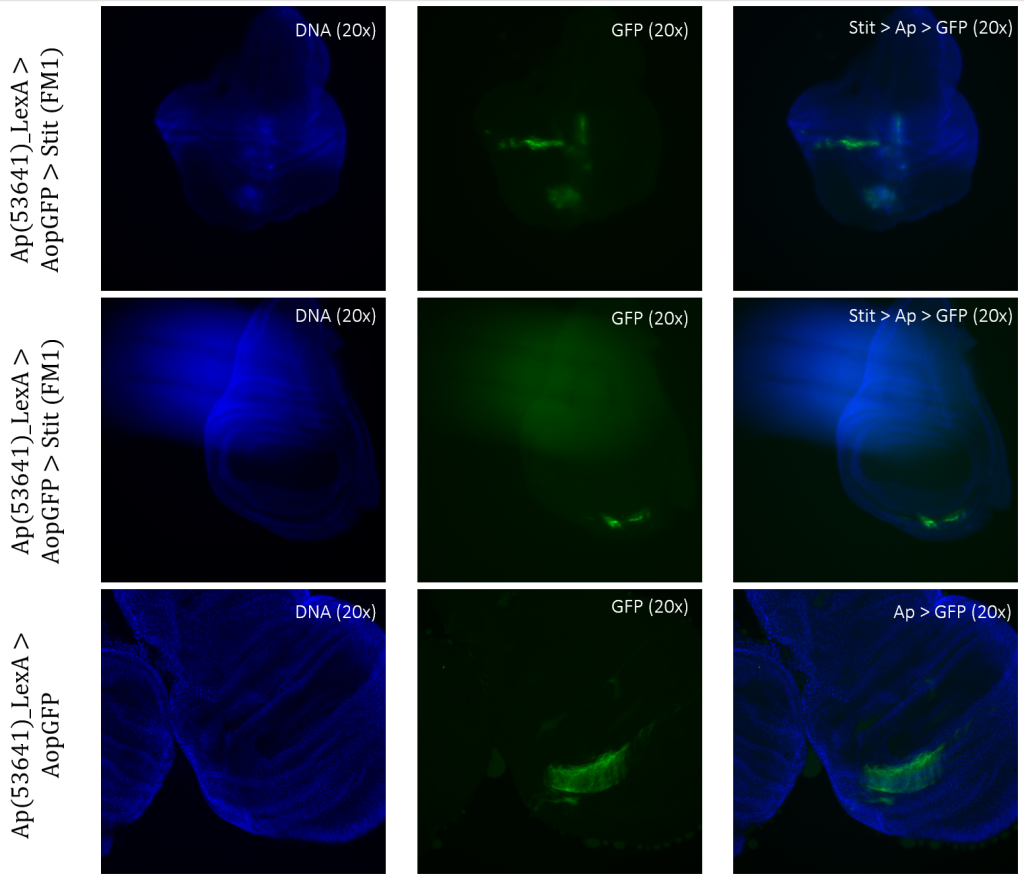
Figure 3.6: Tissue samples from ApLexA promoter for GAL4 expression when exposed to Stit in Drosophila. Wing disk where ApLexA>GFP is shown as fragments within the pouch. Stit-transformation of cells (Stit>Ap>GFP) leads to more diluted fragments of GFP-positive cells within the pouch of the imaginal wing disk. The areas of GFP expression are somewhat consistent with that of the control.
Discussion
Cancer, characterized by uncontrolled growth and spread of abnormal cells, is a major public health concern globally and is one of the leading causes of death (National Cancer Institute, 2023; World Health Organization, 2022). The rise of cancer incidence is attributed to various factors, including changes in lifestyle, exposure to environmental variables, and the aging of the population (National Cancer Institute, 2023). With numerous types of cancer, each having distinct risk factors, symptoms, and therapies. Understanding its causes, preventing its incidence, and finding effective therapies require continuous research and attention.
To investigate the effects of RET/Stit expression on tumour development, we aimed to identify suitable LexA driver lines with restricted expression patterns within the imaginal wing disc of Drosophila. Out of eight LexA drivers tested, five were identified as suitable, and two of them, PtcLexA and ApLexA, were further exposed to Stit for an insight into the effects this might have on the cells. Using the binary system LexA/Aop transcribing GAL4, we identified the expression sites within each genetic line, leading to GFP-positive cells if the gene was present. This way of experimenting along with the use of the two different binary lines is considered to be a decent method for identifying the expression sites based on our results and earlier research within the same scientific field (Rodriguez et al., 2011). Further, the use of balancers has been advantageous as it ensures the whole gene of interest is passed on to the next generation. This mechanism played a crucial role in the project and is a common technique when working with Drosophila gene manipulation (Miller et al., 2019).
Expression of LexA driver lines within the imaginal wing disc of Drosophila
For detection and identification of the expression within the imaginal wing disc of Drosophila, direct crosses of the driver LexA lines to AopGFP were made. The results showed that the lines ApLexA, TrxLexA, PtcLexA, and NubLexA (Figures 4 – 5) had GFP-positive cells within the wing disc of Drosophila, whereas the lines BxLexA, KnLexA, and SalmLexA (Figure 3) were GFP-negative. The GFP-positive cells of Ap-, Trx-, Ptc-, and NubLexA were expressed differently in correlation to the region of expression and total area of expression. NubLexA and PtcLexA had the most fragmented and restricted expressions of the lines (Figure 4 – 5). PtcLexA had a clear fragment in the pouch area and another more elongated expression in the thorax of the wing disc (Figure 4). The NubLexA was highly restricted and showed a lower intensity expression compared to that of the others present in the lower region of the wing pouch (Figure 5). As the NubLexA/CyO was not homozygous (Table 1), only 50% of the larvae had the correct genotype, NubLexA;AopGFP, and the other 50% would be CyO;AopGFP. A higher number of samples to confirm expression was therefore required to confirm GFP-positive cells. An additional control by holding the tube with larvae under a GFP light was also performed (Figure 6). This check confirmed our microscopy dissection results that there were GFP-positive cells in the imaginal wing disc.
TrxLexA had a relatively large expression site compared to that of Nub and Ptc. The GFP-positive cells were restricted to the wing pouch of the imaginal wing disc (Figure 4). Both of the ApLexA lines had high-intensity expression sites. Since these lines were different fragments of the same gene, the expression differed between them. The fragment of Ap(54268)LexA had a larger area of GFP-positive cells than that of Ap(53641) present in both the pouch and the thorax of the imaginal wing disc (Figure 5). Ap(53641) was only present in the pouch of the imaginal wing disc. It is difficult to say whether these fragments have some areas where their expression might overlap as the samples of Ap(53641) were of a later larvae stage. This can be concluded as the pouch has started to fold in this sample (Figure 5).
Exposing selected LexA Driver Lines, PtcLexA and ApLexA, to Oncogene Stit
Exposing PtcLexA and ApLexA to Stit resulted in changed expression sites of GFP-positive cells within the discs when compared to that of the control (Figure 7 – 8). It has been shown through previous studies that Stit is a promoter for migratory of cells (Boekhorst & Friedl, 2016). There are indications of migratory cells of both GFP-positive regions of the PtcLexA and ApLexA when exposed to Stit (Figure 7 – 8). Two different samples of each LexA line exposed to Stit were presented in this study, where the two samples in both cases differed slightly from each other. Stit>Ptc>GFP resulted in more diluted fragments of GFP-positive cells. The GFP-positive cells of the wing pouch had a more elongated pattern reminding of a line rather than a centered fragment like it was in the control (Figure 7). This was only the case for the first sample (Figure 7, column 1); the second sample’s GFP-positive cells in the pouch region were more similar to that of the control, whereas Ptc>GFP. Ptc>GFP also showed a region of GFP-positive cells within the dorsal region of the thorax. This area was also altered when exposed to Stit in the second sample. Here, it looked like the expression was covering a larger area than that of the control (Figure 7, column 2).
Stit>Ap>GFP resulted in more elongated regions of expression, especially within the first sample (Figure 8, column 1). Since this sample was disrupted when mounted, it is not possible to draw a conclusion. However, there are clear indications that the expression changed within the region, resulting in a more long-stacked GFP-positive region. The second sample (Figure 8, column 2) shows two separate fragments of expression in the pouch instead of one completely larger region like in the control, Ap>GFP. Given that the Stit>Ap>GFP wing disc is at an earlier developmental stage than the Ap>GFP wing disc, it is plausible to conclude that the areas of expression are shared, allowing for direct comparisons. (Figure 8). The second sample (Figure 8, column 2) shows signs of movement because there are several small GFP-positive ‘stripes’ elongating from the higher intensity fragment of GFP-positive cells (Figure 8).
Summary of Observations when Exposing LexA Driver Lines to Stit
The images of the samples from both the Stit>Ptc>GFP and Stit>Ap>GFP indicate that there is a movement of cells when exposed to Stit as the expression of GFP-positive cells differs from the control sample results. These findings support the fact that Stit is a promoter for the migration of cells and is further supported in earlier research on Stit (Boekhorst & Friedl, 2016; O’Farrell et al., 2013; Wang et al., 2009). During this experiment, two different Stit lines were used, StitFM1 and Stit FM2.It is not possible to draw comparisons between the two in this case due to the expression with different LexA lines.
Conclusion
In this experiment, we aimed to identify suitable LexA driver lines with restricted expression patterns within the imaginal wing disc of Drosophila. We successfully identified five out of eight LexA drivers to be suitable for comparing the effects of RET/Stit expression in tumour development. Two of the appropriate driver lines, PtcLexA and ApLexA, were further exposed to Stit to gain insight into the effects on the cells. The expression sites within each genetic line were identified using the binary system LexA/Aop transcribing GAL4, which ultimately resulted in GFP-positive cells when the gene is present. Based on our findings and previous research in the same scientific field, this method of experimenting, along with the use of two different binary lines, is regarded as a reasonable method for identifying expression sites (Rodriguez et al., 2011). Our findings suggest that the strength of expression correlates with the penetrance of the tumour phenotype, but additional research is needed due to inconsistencies in expression and a lack of repeated experiments.
Footnotes
[1] Flipping is the act of transferring flies between two tubes. Removing the lids from both tubes - one containing fresh food and the other one with flies - and fast stacking them on top of one another and rotating them. Next, pounding the stacked tubes on the table, forcing the flies down into the new tube. When completed, replace the old lid and discard the empty bottle with its lid still on. [back to text]
How to cite
Solheim, N. (2023). Characterizing Expression Patterns of LexA and LexAop Lines for the Development of a Drosophila Cancer Model. Bikuben 2.
References
Boekhorst, V., & Friedl, P. (2016). Plasticity of Cancer Cell Invasion—Mechanisms and Implications for Therapy. Advances in Cancer Research 132, 209-264. https://doi.org/10.1016/bs.acr.2016.07.005
Eng, C. (1999). RET Proto-Oncogene in the Development of Human Cancer. Journal of Clinical Oncology 17(1), 380-380. DOI: https://doi.org/10.1200/jco.1999.17.1.380
Jennings, B. H. (2011). Drosophila – a versatile model in biology & medicine. Materials Today, 14(5), 190-195. https://doi.org/10.1016/S1369-7021(11)70113-4
Layalle, S., arquier, N., & Léopold, P. (2008). The TOR Pathway Couples Nutrition and Developmental Timing in Drosophila. Developmental Cell, 15(4), 568-577. https://doi.org/10.1016/j.devcel.2008.08.003
Lai, S. L., & Lee, T. (2006). Genetic mosaic with dual binary transcriptional systems in Drosophila. Nature neuroscience, 9(5), 703-709. https://doi.org/10.1038/nn1681
Markow, T. A. (2015). The Natural History of Model Organisms: The secret lives of Drosophila flies. eLife, 4, e06793. https://doi.org/10.7554/eLife.06793
Miller, D. E., Cook, K. R., & Hawley, R. S. (2019). The joy of balancers. PLOS Genetics 15(11), e1008421. https://doi.org/10.1371/journal.pgen.1008421
National Cancer Institute (NCI) at the National Institute of Health (NIH) Malignancy, NCI Dictionaries. (n.d.). Retrieved May 31, 2022, from https://www.cancer.gov/publications/dictionaries/cancer-terms/def/malignancy
O’Farrell, F., Lobert, V., Sneeggen, M., Jain, A., Katheder, N. S., Wenzel, E. M., Schultz, S. W., Tan, K. T., Brech, A., Stenmark, H., & Rusten, T. E. (2017). Class III phosphatidylinositol-3-OH kinase controls epithelial integrity through endosomal LKB1 regulation. Nature Cell Biology 19(12), 1412–1423. https://doi.org/10.1038/ncb3631
Rodríguez, A. V., Didiano, D., & Desplan, C. (2011). Power tools for gene expression and clonal analysis in Drosophila. Nature Methods 9(1), 47–55. https://doi.org/10.1038/nmeth.1800
Schmelzle, T., & Hall, M. N. (2000). TOR, a Central Controller of Cell Growth. Cell 103(2), 253-262. https://doi.org/10.1016/S0092-8674(00)00117-3
Takahashi, M., Kawai, K., & Asai, N. (2020). Roles of the RET Proto-oncogene. Cancer and Development. JMA Journal 3(3), 175-181. https://doi.org/10.31662/jmaj.2020-0021
Wang, S., Tsarouhas, V., Xylourgidis, N., Sabri, N., Tiklová, K., & Nauta, L. et al. (2009). The tyrosine kinase Stitcher activates Grainy head and epidermal wound healing in Drosophila. Nat Cell Biol 11, 890–895 (2009). https://doi.org/10.1038/ncb1898
World Health Organization (2022) Cancer. Retrieved May 2023, from https://www.who.int/news-room/fact-sheets/detail/cancer
About the author
I am incredibly grateful to my supervisor, Fergal O'Farrell, who has provided me with this opportunity. In terms of both my academic and professional goals, this experience has been a turning point, and I am eager to see where it takes me next.”





1 Response
[…] N. (2023). Characterizing Expression Patterns of LexA and LexAop Lines for the Development of a Drosophila Canc…. Bikuben […]